Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
7004: Protein kinases as cancer chemotherapy targets
7004: Protein kinases as cancer chemotherapy targets
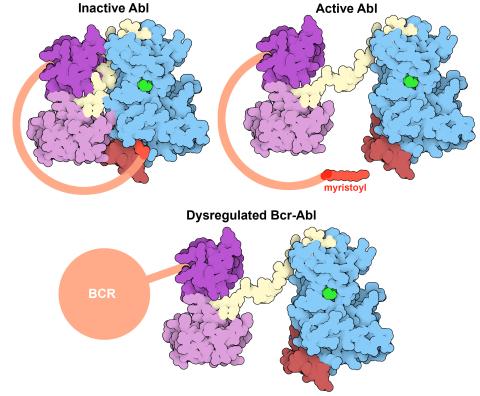
Protein kinases—enzymes that add phosphate groups to molecules—are cancer chemotherapy targets because they play significant roles in almost all aspects of cell function, are tightly regulated, and contribute to the development of cancer and other diseases if any alterations to their regulation occur. Genetic abnormalities affecting the c-Abl tyrosine kinase are linked to chronic myelogenous leukemia, a cancer of immature cells in the bone marrow. In the noncancerous form of the protein, binding of a myristoyl group to the kinase domain inhibits the activity of the protein until it is needed (top left shows the inactive form, top right shows the open and active form). The cancerous variant of the protein, called Bcr-Abl, lacks this autoinhibitory myristoyl group and is continually active (bottom). ATP is shown in green bound in the active site of the kinase.
Find these in the RCSB Protein Data Bank: c-Abl tyrosine kinase and regulatory domains (PDB entry 1OPL) and F-actin binding domain (PDB entry 1ZZP).
Find these in the RCSB Protein Data Bank: c-Abl tyrosine kinase and regulatory domains (PDB entry 1OPL) and F-actin binding domain (PDB entry 1ZZP).
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media

2475: Chromosome fiber 01
2475: Chromosome fiber 01
This microscopic image shows a chromatin fiber--a DNA molecule bound to naturally occurring proteins.
Marc Green and Susan Forsburg, University of Southern California
View Media

3333: Polarized cells- 02
3333: Polarized cells- 02
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3-/- fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red) and DAPI to visualize the nucleus (blue). In the absence of functional Arp2/3 complex, ARPC3-/- fibroblast cells' leading edge morphology is significantly altered with filopodia-like structures. Related to images 3328, 3329, 3330, 3331, and 3332.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media

2503: Focal adhesions (with labels)
2503: Focal adhesions (with labels)
Cells walk along body surfaces via tiny "feet," called focal adhesions, that connect with the extracellular matrix. See image 2502 for an unlabeled version of this illustration.
Crabtree + Company
View Media

1282: Lysosomes
1282: Lysosomes
Lysosomes have powerful enzymes and acids to digest and recycle cell materials.
Judith Stoffer
View Media

3253: Pulsating response to stress in bacteria
3253: Pulsating response to stress in bacteria
By attaching fluorescent proteins to the genetic circuit responsible for B. subtilis's stress response, researchers can observe the cells' pulses as green flashes. In response to a stressful environment like one lacking food, B. subtilis activates a large set of genes that help it respond to the hardship. Instead of leaving those genes on as previously thought, researchers discovered that the bacteria flip the genes on and off, increasing the frequency of these pulses with increasing stress. See entry 3254 for the related video.
Michael Elowitz, Caltech University
View Media

1241: Borrelia burgdorferi
1241: Borrelia burgdorferi
Borrelia burgdorferi is a spirochete, a class of long, slender bacteria that typically take on a coiled shape. Infection with this bacterium causes Lyme disease.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media

5793: Mouse retina
5793: Mouse retina
What looks like the gossamer wings of a butterfly is actually the retina of a mouse, delicately snipped to lay flat and sparkling with fluorescent molecules. The image is from a research project investigating the promise of gene therapy for glaucoma. It was created at an NIGMS-funded advanced microscopy facility that develops technology for imaging across many scales, from whole organisms to cells to individual molecules.
The ability to obtain high-resolution imaging of tissue as large as whole mouse retinas was made possible by a technique called large-scale mosaic confocal microscopy, which was pioneered by the NIGMS-funded National Center for Microscopy and Imaging Research. The technique is similar to Google Earth in that it computationally stitches together many small, high-resolution images.
The ability to obtain high-resolution imaging of tissue as large as whole mouse retinas was made possible by a technique called large-scale mosaic confocal microscopy, which was pioneered by the NIGMS-funded National Center for Microscopy and Imaging Research. The technique is similar to Google Earth in that it computationally stitches together many small, high-resolution images.
Tom Deerinck and Keunyoung (“Christine”) Kim, NCMIR
View Media

6752: Petri dish
6752: Petri dish
The white circle in this image is a Petri dish, named for its inventor, Julius Richard Petri. These dishes are one of the most common pieces of equipment in biology labs, where researchers use them to grow cells.
H. Robert Horvitz and Dipon Ghosh, Massachusetts Institute of Technology.
View Media

5754: Zebrafish pigment cell
5754: Zebrafish pigment cell
Pigment cells are cells that give skin its color. In fishes and amphibians, like frogs and salamanders, pigment cells are responsible for the characteristic skin patterns that help these organisms to blend into their surroundings or attract mates. The pigment cells are derived from neural crest cells, which are cells originating from the neural tube in the early embryo. Investigating pigment cell formation and migration in animals helps answer important fundamental questions about the factors that control pigmentation in the skin of animals, including humans. This image shows a pigment cell from zebrafish at high resolution. Related to images 5755, 5756, 5757 and 5758.
David Parichy, University of Washington
View Media

6802: Antibiotic-surviving bacteria
6802: Antibiotic-surviving bacteria
Colonies of bacteria growing despite high concentrations of antibiotics. These colonies are visible both by eye, as seen on the left, and by bioluminescence imaging, as seen on the right. The bioluminescent color indicates the metabolic activity of these bacteria, with their red centers indicating high metabolism.
More information about the research that produced this image can be found in the Antimicrobial Agents and Chemotherapy paper “Novel aminoglycoside-tolerant phoenix colony variants of Pseudomonas aeruginosa” by Sindeldecker et al.
More information about the research that produced this image can be found in the Antimicrobial Agents and Chemotherapy paper “Novel aminoglycoside-tolerant phoenix colony variants of Pseudomonas aeruginosa” by Sindeldecker et al.
Paul Stoodley, The Ohio State University.
View Media

2325: Multicolor STORM
2325: Multicolor STORM
In 2006, scientists developed an optical microscopy technique enabling them to clearly see individual molecules within cells. In 2007, they took the technique, abbreviated STORM, a step further. They identified multicolored probes that let them peer into cells and clearly see multiple cellular components at the same time, such as these microtubules (green) and small hollows called clathrin-coated pits (red). Unlike conventional methods, the multicolor STORM technique produces a crisp and high resolution picture. A sharper view of how cellular components interact will likely help scientists answer some longstanding questions about cell biology.
Xiaowei Zhuang, Harvard University
View Media

2690: Dolly the sheep
2690: Dolly the sheep
Scientists in Scotland were the first to clone an animal, this sheep named Dolly. She later gave birth to Bonnie, the lamb next to her.
View Media

3549: TonB protein in gram-negative bacteria
3549: TonB protein in gram-negative bacteria
The green in this image highlights a protein called TonB, which is produced by many gram-negative bacteria, including those that cause typhoid fever, meningitis and dysentery. TonB lets bacteria take up iron from the host's body, which they need to survive. More information about the research behind this image can be found in a Biomedical Beat Blog posting from August 2013.
Phillip Klebba, Kansas State University
View Media
3265: Microfluidic chip
3265: Microfluidic chip
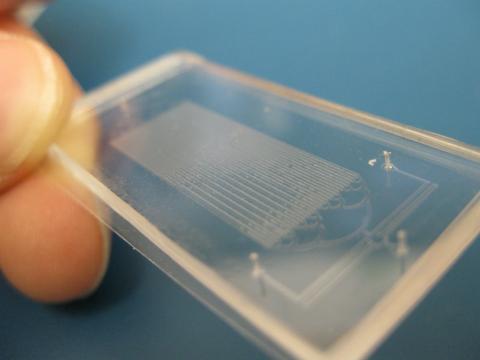
Microfluidic chips have many uses in biology labs. The one shown here was used by bioengineers to study bacteria, allowing the researchers to synchronize their fluorescing so they would blink in unison. Related to images 3266 and 3268. From a UC San Diego news release, "Researchers create living 'neon signs' composed of millions of glowing bacteria."
Jeff Hasty Lab, UC San Diego
View Media

2743: Molecular interactions
2743: Molecular interactions
This network map shows molecular interactions (yellow) associated with a congenital condition that causes heart arrhythmias and the targets for drugs that alter these interactions (red and blue).
Ravi Iyengar, Mount Sinai School of Medicine
View Media

2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
This image represents the structure of TrpRS, a novel member of the tryptophanyl tRNA-synthetase family of enzymes. By helping to link the amino acid tryptophan to a tRNA molecule, TrpRS primes the amino acid for use in protein synthesis. A cluster of iron and sulfur atoms (orange and red spheres) was unexpectedly found in the anti-codon domain, a key part of the molecule, and appears to be critical for the function of the enzyme. TrpRS was discovered in Thermotoga maritima, a rod-shaped bacterium that flourishes in high temperatures.
View Media

2381: dUTP pyrophosphatase from M. tuberculosis
2381: dUTP pyrophosphatase from M. tuberculosis
Model of an enzyme, dUTP pyrophosphatase, from Mycobacterium tuberculosis. Drugs targeted to this enzyme might inhibit the replication of the bacterium that causes most cases of tuberculosis.
Mycobacterium Tuberculosis Center, PSI
View Media

2561: Histones in chromatin (with labels)
2561: Histones in chromatin (with labels)
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2560 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
2368: Mounting of protein crystals
2368: Mounting of protein crystals
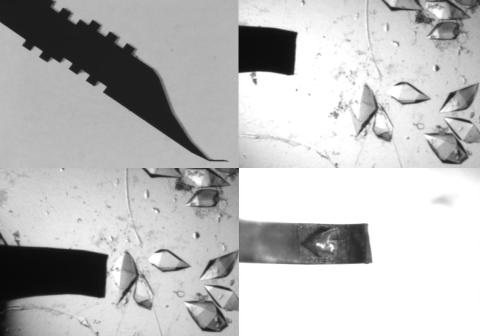
Automated methods using micromachined silicon are used at the Northeast Collaboratory for Structural Genomics to mount protein crystals for X-ray crystallography.
The Northeast Collaboratory for Structural Genomics
View Media
2324: Movements of myosin
2324: Movements of myosin
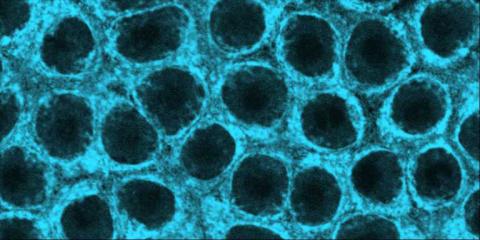
Inside the fertilized egg cell of a fruit fly, we see a type of myosin (related to the protein that helps muscles contract) made to glow by attaching a fluorescent protein. After fertilization, the myosin proteins are distributed relatively evenly near the surface of the embryo. The proteins temporarily vanish each time the cells' nuclei--initially buried deep in the cytoplasm--divide. When the multiplying nuclei move to the surface, they shift the myosin, producing darkened holes. The glowing myosin proteins then gather, contract, and start separating the nuclei into their own compartments.
Victoria Foe, University of Washington
View Media

2604: Induced stem cells from adult skin 02
2604: Induced stem cells from adult skin 02
These cells are induced stem cells made from human adult skin cells that were genetically reprogrammed to mimic embryonic stem cells. The induced stem cells were made potentially safer by removing the introduced genes and the viral vector used to ferry genes into the cells, a loop of DNA called a plasmid. The work was accomplished by geneticist Junying Yu in the laboratory of James Thomson, a University of Wisconsin-Madison School of Medicine and Public Health professor and the director of regenerative biology for the Morgridge Institute for Research.
James Thomson, University of Wisconsin-Madison
View Media

6992: Molecular view of glutamatergic synapse
6992: Molecular view of glutamatergic synapse
This illustration highlights spherical pre-synaptic vesicles that carry the neurotransmitter glutamate. The presynaptic and postsynaptic membranes are shown with proteins relevant for transmitting and modulating the neuronal signal.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media

5729: Assembly of the HIV capsid
5729: Assembly of the HIV capsid
The HIV capsid is a pear-shaped structure that is made of proteins the virus needs to mature and become infective. The capsid is inside the virus and delivers the virus' genetic information into a human cell. To better understand how the HIV capsid does this feat, scientists have used computer programs to simulate its assembly. This image shows a series of snapshots of the steps that grow the HIV capsid. A model of a complete capsid is shown on the far right of the image for comparison; the green, blue and red colors indicate different configurations of the capsid protein that make up the capsid “shell.” The bar in the left corner represents a length of 20 nanometers, which is less than a tenth the size of the smallest bacterium. Computer models like this also may be used to reconstruct the assembly of the capsids of other important viruses, such as Ebola or the Zika virus. The studies reporting this research were published in Nature Communications and Nature. To learn more about how researchers used computer simulations to track the assembly of the HIV capsid, see this press release from the University of Chicago.
John Grime and Gregory Voth, The University of Chicago
View Media

3771: Molecular model of freshly made Rous sarcoma virus (RSV)
3771: Molecular model of freshly made Rous sarcoma virus (RSV)
Viruses have been the foes of animals and other organisms for time immemorial. For almost as long, they've stayed well hidden from view because they are so tiny (they aren't even cells, so scientists call the individual virus a "particle"). This image shows a molecular model of a particle of the Rous sarcoma virus (RSV), a virus that infects and sometimes causes cancer in chickens. In the background is a photo of red blood cells. The particle shown is "immature" (not yet capable of infecting new cells) because it has just budded from an infected chicken cell and entered the bird's bloodstream. The outer shell of the immature virus is made up of a regular assembly of large proteins (shown in red) that are linked together with short protein molecules called peptides (green). This outer shell covers and protects the proteins (blue) that form the inner shell of the particle. But as you can see, the protective armor of the immature virus contains gaping holes. As the particle matures, the short peptides are removed and the large proteins rearrange, fusing together into a solid sphere capable of infecting new cells. While still immature, the particle is vulnerable to drugs that block its development. Knowing the structure of the immature particle may help scientists develop better medications against RSV and similar viruses in humans. Scientists used sophisticated computational tools to reconstruct the RSV atomic structure by crunching various data on the RSV proteins to simulate the entire structure of immature RSV.
Boon Chong Goh, University of Illinois at Urbana-Champaign
View Media

3426: Regeneration of Mouse Ears
3426: Regeneration of Mouse Ears
Normal mice, like the B6 breed pictured on the left, develop scars when their ears are pierced. The Murphy Roths Large (MRL) mice pictured on the right can grow back lost ear tissue thanks to an inactive version of the p21 gene. When researchers knocked out that same gene in other mouse breeds, their ears also healed completely without scarring. Journal Article: Clark, L.D., Clark, R.K. and Heber-Katz, E. 1998. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol 88: 35-45.
Ellen Heber-Katz, The Wistar Institute
View Media

1275: Golgi
1275: Golgi
The Golgi complex, also called the Golgi apparatus or, simply, the Golgi. This organelle receives newly made proteins and lipids from the ER, puts the finishing touches on them, addresses them, and sends them to their final destinations.
Judith Stoffer
View Media

1285: Lipid raft
1285: Lipid raft
Researchers have learned much of what they know about membranes by constructing artificial membranes in the laboratory. In artificial membranes, different lipids separate from each other based on their physical properties, forming small islands called lipid rafts.
Judith Stoffer
View Media

2607: Mouse embryo showing Smad4 protein
2607: Mouse embryo showing Smad4 protein
This eerily glowing blob isn't an alien or a creature from the deep sea--it's a mouse embryo just eight and a half days old. The green shell and core show a protein called Smad4. In the center, Smad4 is telling certain cells to begin forming the mouse's liver and pancreas. Researchers identified a trio of signaling pathways that help switch on Smad4-making genes, starting immature cells on the path to becoming organs. The research could help biologists learn how to grow human liver and pancreas tissue for research, drug testing and regenerative medicine. In addition to NIGMS, NIH's National Institute of Diabetes and Digestive and Kidney Diseases also supported this work.
Kenneth Zaret, Fox Chase Cancer Center
View Media

5762: Panorama view of golden mitochondria
5762: Panorama view of golden mitochondria
Mitochondria are the powerhouses of the cells, generating the energy the cells need to do their tasks and to stay alive. Researchers have studied mitochondria for some time because when these cell organelles don't work as well as they should, several diseases develop. In this photograph of cow cells taken with a microscope, the mitochondria were stained in bright yellow to visualize them in the cell. The large blue dots are the cell nuclei and the gray web is the cytoskeleton of the cells.
Torsten Wittmann, University of California, San Francisco
View Media

2540: Chromosome inside nucleus (with labels)
2540: Chromosome inside nucleus (with labels)
The long, stringy DNA that makes up genes is spooled within chromosomes inside the nucleus of a cell. (Note that a gene would actually be a much longer stretch of DNA than what is shown here.) See image 2539 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media

2556: Dicer generates microRNAs
2556: Dicer generates microRNAs
The enzyme Dicer generates microRNAs by chopping larger RNA molecules into tiny Velcro®-like pieces. MicroRNAs stick to mRNA molecules and prevent the mRNAs from being made into proteins. See image 2557 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
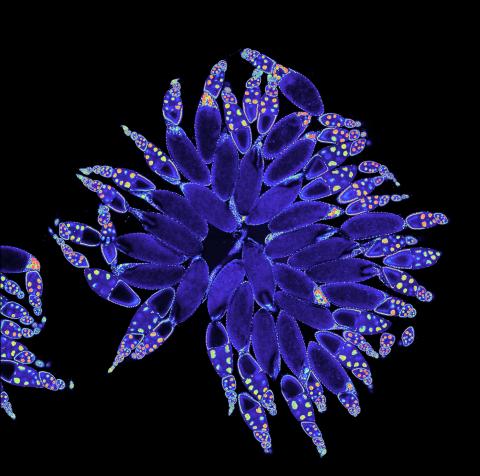
3607: Fruit fly ovary
3607: Fruit fly ovary
A fruit fly ovary, shown here, contains as many as 20 eggs. Fruit flies are not merely tiny insects that buzz around overripe fruit—they are a venerable scientific tool. Research on the flies has shed light on many aspects of human biology, including biological rhythms, learning, memory, and neurodegenerative diseases. Another reason fruit flies are so useful in a lab (and so successful in fruit bowls) is that they reproduce rapidly. About three generations can be studied in a single month.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Denise Montell, Johns Hopkins University and University of California, Santa Barbara
View Media

1330: Mitosis - prophase
1330: Mitosis - prophase
A cell in prophase, near the start of mitosis: In the nucleus, chromosomes condense and become visible. In the cytoplasm, the spindle forms. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media

7011: Hawaiian bobtail squid
7011: Hawaiian bobtail squid
An adult Hawaiian bobtail squid, Euprymna scolopes, swimming next to a submerged hand.
Related to image 7010 and video 7012.
Related to image 7010 and video 7012.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media

5752: Genetically identical mycobacteria respond differently to antibiotic 2
5752: Genetically identical mycobacteria respond differently to antibiotic 2
Antibiotic resistance in microbes is a serious health concern. So researchers have turned their attention to how bacteria undo the action of some antibiotics. Here, scientists set out to find the conditions that help individual bacterial cells survive in the presence of the antibiotic rifampicin. The research team used Mycobacterium smegmatis, a more harmless relative of Mycobacterium tuberculosis, which infects the lung and other organs to cause serious disease.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
Bree Aldridge, Tufts University
View Media

3656: Fruit fly ovary_2
3656: Fruit fly ovary_2
A fruit fly ovary, shown here, contains as many as 20 eggs. Fruit flies are not merely tiny insects that buzz around overripe fruit--they are a venerable scientific tool. Research on the flies has shed light on many aspects of human biology, including biological rhythms, learning, memory and neurodegenerative diseases. Another reason fruit flies are so useful in a lab (and so successful in fruit bowls) is that they reproduce rapidly. About three generations can be studied in a single month. Related to image 3607.
Denise Montell, University of California, Santa Barbara
View Media

1334: Aging book of life
1334: Aging book of life
Damage to each person's genome, often called the "Book of Life," accumulates with time. Such DNA mutations arise from errors in the DNA copying process, as well as from external sources, such as sunlight and cigarette smoke. DNA mutations are known to cause cancer and also may contribute to cellular aging.
Judith Stoffer
View Media

3670: DNA and actin in cultured fibroblast cells
3670: DNA and actin in cultured fibroblast cells
DNA (blue) and actin (red) in cultured fibroblast cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

6781: Video of Calling Cards in a mouse brain
6781: Video of Calling Cards in a mouse brain
The green spots in this mouse brain are cells labeled with Calling Cards, a technology that records molecular events in brain cells as they mature. Understanding these processes during healthy development can guide further research into what goes wrong in cases of neuropsychiatric disorders. Also fluorescently labeled in this video are neurons (red) and nuclei (blue). Calling Cards and its application are described in the Cell paper “Self-Reporting Transposons Enable Simultaneous Readout of Gene Expression and Transcription Factor Binding in Single Cells” by Moudgil et al.; and the Proceedings of the National Academy of Sciences paper “A viral toolkit for recording transcription factor–DNA interactions in live mouse tissues” by Cammack et al. This video was created for the NIH Director’s Blog post The Amazing Brain: Tracking Molecular Events with Calling Cards.
Related to image 6780.
Related to image 6780.
NIH Director's Blog
View Media

6755: Honeybee brain
6755: Honeybee brain
Insect brains, like the honeybee brain shown here, are very different in shape from human brains. Despite that, bee and human brains have a lot in common, including many of the genes and neurochemicals they rely on in order to function. The bright-green spots in this image indicate the presence of tyrosine hydroxylase, an enzyme that allows the brain to produce dopamine. Dopamine is involved in many important functions, such as the ability to experience pleasure. This image was captured using confocal microscopy.
Gene Robinson, University of Illinois at Urbana-Champaign.
View Media

3491: Kinesin moves cellular cargo
3491: Kinesin moves cellular cargo
A protein called kinesin (blue) is in charge of moving cargo around inside cells and helping them divide. It's powered by biological fuel called ATP (bright yellow) as it scoots along tube-like cellular tracks called microtubules (gray).
Charles Sindelar, Yale University
View Media

6848: Himastatin
6848: Himastatin
A model of the molecule himastatin, which was first isolated from the bacterium Streptomyces himastatinicus. Himastatin shows antibiotic activity. The researchers who created this image developed a new, more concise way to synthesize himastatin so it can be studied more easily.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6850 and video 6851.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6850 and video 6851.
Mohammad Movassaghi, Massachusetts Institute of Technology.
View Media

3756: Protective membrane and membrane proteins of the dengue virus visualized with cryo-EM
3756: Protective membrane and membrane proteins of the dengue virus visualized with cryo-EM
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. You can watch a rotating view of the dengue virus surface structure in video 3748.
Hong Zhou, UCLA
View Media

6344: Drosophila
6344: Drosophila
Two adult fruit flies (Drosophila)
Dr. Vicki Losick, MDI Biological Laboratory, www.mdibl.org
View Media
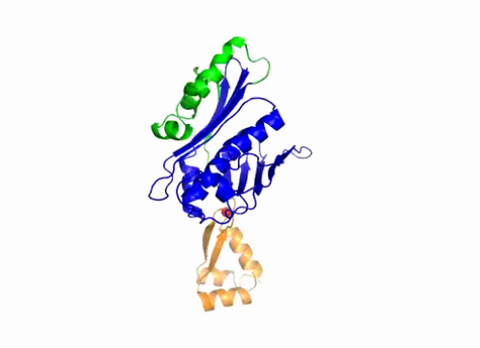
3402: Hsp33 Heat Shock Protein Inactive to Active
3402: Hsp33 Heat Shock Protein Inactive to Active
When the heat shock protein hsp33 is folded, it is inactive and contains a zinc ion, stabilizing the redox sensitive domain (orange). In the presence of an environmental stressor, the protein releases the zinc ion, which leads to the unfolding of the redox domain. This unfolding causes the chaperone to activate by reaching out its "arm" (green) to protect other proteins.
Dana Reichmann, University of Michigan
View Media

6768: Rhodopsin bound to visual arrestin
6768: Rhodopsin bound to visual arrestin
Rhodopsin is a pigment in the rod cells of the retina (back of the eye). It is extremely light-sensitive, supporting vision in low-light conditions. Here, it is attached to arrestin, a protein that sends signals in the body. This structure was determined using an X-ray free electron laser.
Protein Data Bank.
View Media

6811: Fruit fly egg chamber
6811: Fruit fly egg chamber
A fruit fly (Drosophila melanogaster) egg chamber with microtubules shown in green and actin filaments shown in red. Egg chambers are multicellular structures in fruit flies ovaries that each give rise to a single egg. Microtubules and actin filaments give the chambers structure and shape. This image was captured using a confocal microscope.
More information on the research that produced this image can be found in the Current Biology paper "Gatekeeper function for Short stop at the ring canals of the Drosophila ovary" by Lu et al.
More information on the research that produced this image can be found in the Current Biology paper "Gatekeeper function for Short stop at the ring canals of the Drosophila ovary" by Lu et al.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media

6534: Mosaicism in C. elegans (White Background)
6534: Mosaicism in C. elegans (White Background)
In the worm C. elegans, double-stranded RNA made in neurons can silence matching genes in a variety of cell types through the transport of RNA between cells. The head region of three worms that were genetically modified to express a fluorescent protein were imaged and the images were color-coded based on depth. The worm on the left lacks neuronal double-stranded RNA and thus every cell is fluorescent. In the middle worm, the expression of the fluorescent protein is silenced by neuronal double-stranded RNA and thus most cells are not fluorescent. The worm on the right lacks an enzyme that amplifies RNA for silencing. Surprisingly, the identities of the cells that depend on this enzyme for gene silencing are unpredictable. As a result, worms of identical genotype are nevertheless random mosaics for how the function of gene silencing is carried out. For more, see journal article and press release. Related to image 6532.
Snusha Ravikumar, Ph.D., University of Maryland, College Park, and Antony M. Jose, Ph.D., University of Maryland, College Park
View Media

7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
The beating of cilia on the outside of the Hawaiian bobtail squid’s light organ concentrates Vibrio fischeri cells (green) present in the seawater into aggregates near the pore-containing tissue (red). From there, the bacterial cells (~2 mm) swim to the pores and migrate through a bottleneck into the interior crypts where a population of symbionts grow and remain for the life of the host. This image was taken using confocal fluorescence microscopy.
Related to images 7016, 7017, 7018, and 7020.
Related to images 7016, 7017, 7018, and 7020.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
